Andrés Pérez-Figueroa
A biologist that prefers dealing with code bugs rather than live bugs.
Interdisciplinary Centre of Marine and Environmental Research (CIIMAR)
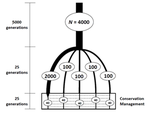
Hi! I’m Andrés Pérez-Figueroa, a postdoctoral researcher in the Interdisciplinary Centre of Marine and Environmental Research (CIIMAR) in Portugal. My current research here aims to decipher the adaptive forces in cancer evolution acting at the ultimate level of resolution, the single-cell. My background is related to conservation and population genetics as a computational biologist. I can write code (not very well) to simulate and analyze biological data for understanding its evolutionary implications.
Interests
- population genetics
- cancer
- genetic diversity
- conservation genetics
- epigenetic differentiation
- quantitative genetics
- data science
Education
-
PhD in Genetics, 2005
University of Vigo
-
MSc in Biology, 2002
University of Vigo
-
BSc in Biology, 2000
University of Vigo